Learn how to optimize your Python data pipeline code to run faster with our high-performance Python guide for data engineers.

The following article is part of a series on Python for data engineering aimed at helping data engineers, data scientists, data analysts, Machine Learning engineers, or others who are new to Python master the basics. To date this beginners guide consists of:
- Part 1: Python Packages: a Primer for Data People (part 1 of 2), explored the basics of Python modules, Python packages and how to import modules into your own projects.
- Part 2: Python Packages: a Primer for Data People (part 2 of 2), covered dependency management and virtual environments.
- Part 3: Best Practices in Structuring Python Projects, covered 9 best practices and examples on structuring your projects.
- Part 4: From Python Projects to Dagster Pipelines, we explore setting up a Dagster project, and the key concept of Data Assets.
- Part 5: Environment Variables in Python, we cover the importance of environment variables and how to use them.
- Part 6: Type Hinting, or how type hints reduce errors.
- Part 7: Factory Patterns, or learning design patterns, which are reusable solutions to common problems in software design.
- Part 8: Write-Audit-Publish in data pipelines a design pattern frequently used in ETL to ensure data quality and reliability.
- Part 9: CI/CD and Data Pipeline Atomation (with Git), learn to automate data pipelines and deployments with Git.
- Part 10: High-performance Python for Data Engineering, learn how to code data pipelines in Python for performance.
- Part 11: Breaking Packages in Python, in which we explore the sharp edges of Python’s system of imports, modules, and packages.
Sign up for our newsletter to get all the updates! And if you enjoyed this guide check out our data engineering glossary, complete with Python code examples.
Successful data pipelines do not just rest on writing functional code. As datasets grow larger and processing needs become more complex, we must ensure that our Python code is also efficient.
But what does high-performance code look like?
It's a blend of speed, efficiency, and scalability.
In this article, we’ll discuss the art of optimizing code for better performance. We’ll share practical, hands-on advice to help data engineers avoid common pitfalls, recognize the patterns that slow things down, and improve Python code efficiency across data engineering platforms and tools.
What is optimization and performance? A definition in data engineering
In data engineering, Python code optimization and improving performance means refining code to achieve the most efficient execution with the least resource overhead while ensuring seamless data flow and processing. The techniques in this article would apply to code in another language (such as Rust or Java), at least conceptually.
Table of contents
- Optimization and performance
- Performance and data storage
- Beyond Pandas
- Python types and specialized structures
- Towards more efficient in-memoryPython pipelines
Optimization and performance of Python code
If you’re new to coding, thinking about how to improve Python performance may sound abstract. But it boils down to making your code run faster and use fewer resources like less memory and CPU power.
Let's say you've engineered a data pipeline to cleanse and organize a hefty dataset for further analysis, but it takes an entire hour to process the data every time you run or test the pipeline. Multiply this by tens or hundreds of pipelines across the company, and it’s clear how these inefficiencies compound, too.
You could trim this processing time to just a few minutes with some relatively easy optimizations. Optimized code isn't just faster. It's also more cost-effective, making the most of every computational cycle and dollar spent.
Python performance and data storage
Before diving into technical examples, let’s discuss data storage and why better Python performance matters.
Data storage 101
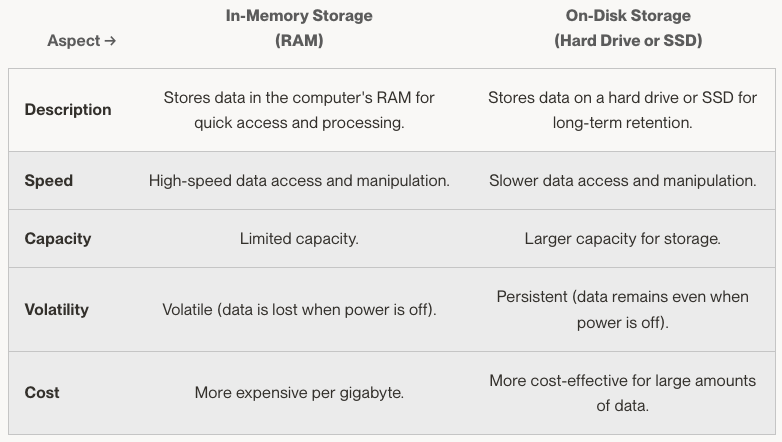
Data can be stored in two primary places: in memory (RAM - Random Access Memory) or on disk (your computer's hard drive or an SSD - Solid State Drive).
- In-Memory Storage (RAM):
When we talk about in-memory storage, we refer to storing in the computer's RAM. This is the memory your computer uses to hold data being used or processed at any given moment. It's like the desk you work on, holding the papers you are reading or writing on.
- On-Disk Storage (Hard Drive or SSD):
On-disk storage saves data on your computer's hard drive or an SSD. This is where data is stored for the long term, even when your computer is turned off. It's like a filing cabinet where you store documents that you don't need right now but will need later.
Performance trade-offs
The choice between in-memory and on-disk storage can significantly impact the application's performance and the efficiency of your processing tasks.
Understanding the trade-offs and implications of each storage type becomes extremely relevant in many common data engineering scenarios. We’ll take a look at a common one in the next section.
Many practitioners use Python libraries like Pandas and NumPy because their built-in functions make handling data a breeze, but it can lead to performance issues and ramp up costs.
One typical trap many early data engineers fall into is using Pandas for everything. While Pandas makes it easy to wrangle and analyze data, using it for simpler tasks or very large datasets can lead to slower execution and higher RAM usage. It’s like using a sledgehammer to crack a nut—it gets the job done, but it's overkill and not the most efficient way to tackle the task at hand.
Let’s dive deeper into Pandas’ performance issues because it’ll help illustrate how you can apply the same principles to other libraries. Pandas isn’t the most efficient choice for two reasons.
- Pandas' overhead
Pandas is designed to simplify complex data operations, but this ease of use comes with a trade-off in performance, especially for simple tasks or operations. Even the latest version of Pandas can introduce additional processing steps, which could lead to slower execution compared to using basic Python data structures like lists or dictionaries.
For example, a simple operation like summing a series of numbers (the list in square brackets) could be executed faster with native Python compared to Pandas, mainly when dealing with a small dataset:
Summing the series of numbers using native Python with the sum() function is straightforward. On the other hand, to perform the same operation using Pandas, you need to first convert the list to a Pandas Series and then use the sum() method of the Series object. This additional step of converting the list to a Series introduces extra overhead, which, while not significant for this small dataset, can add up when performing many such simple operations.
If you only need to perform a basic operation on a dataset, using a Pandas DataFrame might be overkill and could make your code harder to understand compared to using a simple Python list or dictionary.
- Memory consumption
Pandas often loads the entire dataset into RAM for fast data access and manipulation. While this is beneficial for medium-sized datasets, it can become a problem when dealing with very large ones that exceed the available RAM.
In such scenarios, using Pandas can lead to RAM exhaustion, causing your program to crash or run extremely slowly.
One way to get around this is to use libraries designed for out-of-core or distributed computing:
- Dask: Dask is similar to Pandas but can handle larger-than-memory datasets by breaking them down into smaller chunks, processing them in parallel, and even distributing them across multiple machines.
- Vaex: Vaex is another library that can handle large datasets efficiently by performing operations lazily and utilizing memory-mapped files.
Another option is to use on-disk storage solutions, especially when dealing with massive datasets or when persistence is required:
- SQL Databases: Utilize SQL databases like PostgreSQL or SQLite to manage and query your data on-disk
- NoSQL Databases: For unstructured or semi-structured data, NoSQL databases like MongoDB can be a good choice
- Apache Parquet: It's a columnar storage file format optimized for use with big data processing frameworks. It's efficient for both storage and processing, especially when working with complex nested data structures
It’s probably most likely that you’ll use a hybrid approach: use on-disk storage for long-term data retention and in-memory processing for active data manipulation and analysis.
Python types and specialized structures
Selecting the right data types in Python is crucial for writing high-performance code, especially in applications where speed and RAM efficiency are essential. Python, being a dynamically-typed language, allows for flexibility in choosing data types, but this flexibility can lead to inefficiencies if not used judiciously.
Types in the Python standard library
Furthermore, the appropriate choice of data type can affect the algorithmic complexity and, consequently, the performance of the code. For instance, choosing a Python dictionary (a hash table) for frequent lookup operations is much faster than using a list, as dictionaries have average-case constant time complexity for lookups, whereas lists have linear time complexity.
More advanced concepts like the Python generator object can further help you improve code performance. Generator objects and a special type of iterable, like a list or a tuple, but are particularly memory-efficient when dealing with large datasets or streams of data.
Looking to specialized structures
One key consideration is choosing between standard Python data structures like lists and specialized structures like NumPy arrays. Python lists are versatile and easy to use but inefficient for numerical operations on large data sets. They can store different types of objects, which adds overhead in terms of RAM and processing. For instance, when a list contains objects of different types, Python must store type information for each element and perform type checking during runtime, which can slow down computation.
On the other hand, NumPy arrays, often used for numerical computing in Python, and help improve performance of operations on large files of homogeneous data. These arrays use contiguous blocks of memory, making access and manipulation of array elements more efficient. They also support vectorized operations (which cannot be done with a standard library), allowing for complex computations to be executed on entire arrays without the need for explicit loops in Python. This not only leads to more concise and readable code but also significantly enhances performance due to optimized, low-level C implementations for mathematical operations.
In summary, the thoughtful selection of data types is vital in Python for optimizing both speed and memory usage. While Python's flexibility and ease of use are advantageous, leveraging specialized data types like NumPy arrays for numerical operations and making smart choices between different native data structures can significantly improve the performance of Python code. This is especially true in data-intensive and computationally demanding applications, such as scientific computing, data analysis, and machine learning.
Python code optimization and global variables
Global variables in Python are those defined outside of a function and which can be accessed from any part of the program, as explored earlier in the guide. However, their use often sparks debate regarding best practices in coding, especially concerning code optimization and maintainability.
From a performance optimization perspective, global variables can sometimes offer benefits. They can be used to cache values that are expensive to compute and are required by multiple functions, potentially reducing the need for redundant calculations. This can lead to performance improvements in certain scenarios.
Nevertheless, there are many scenarios where we should avoid global variables:
- It can lead to code that is hard to debug and maintain as global variables can be modified by any part of the program.
- Using global variables can lead to issues with code scalability and readability. They make it harder to understand the interactions between different parts of the code, and more challenging to test or implement changes without unintended side effects.
- From a design perspective, overusing global variables often indicates a lack of encapsulation, affecting code modularity and reusability.
So, while global variables can be used for optimizing performance in specific cases, they should be used sparingly and judiciously. It's often better to consider alternative approaches, such as function arguments or class attributes, which can provide a cleaner code that is easier to test.
Towards more efficient in-memory Python pipelines
Let's now turn to some practical advice on how Python code can be optimized to efficiently process data in-memory.
Vectorized data transformations
Always opt for vectorized operations over traditional loops. Vectorized operations refer to performing operations on entire arrays or collections of data at once rather than iterating through each element individually.
In Python, the NumPy library is well-known for providing vectorized operations. Here, we create a NumPy array data_array with some initial values and then perform a vectorized multiplication operation on the entire array.
Efficient data loading
For datasets that can fit into available RAM, Pandas provides efficient tools for data manipulation:
Chunking
When dealing with large datasets, however, it can be impractical to load them into RAM all at once. As we discussed earlier about storage and performance trade-offs, loading these into RAM can be very slow or even cause your program to crash.
Instead, data engineers often use techniques like streaming, chunking, or using databases that allow for processing data in smaller, manageable pieces. Pandas provides a method to read data in chunks:
In this example, the large_dataset.csv file is read and processed in chunks. The size of each chunk is defined by the chunk_size variable. The sum of the some_column column is computed chunk by chunk, and the results are aggregated to get the total sum.
Lazy evaluation
Lazy evaluation is a programming concept where the execution of expressions is delayed until the value of the expression is actually needed. Instead of immediately performing computations when you define them, they're "stored" or "remembered" by the system. Only when you specifically ask for a result (like calling a function), the actual work is done. Pandas does eager evaluation: when you run an operation, the result is computed immediately.
Libraries like Vaex or Polars in Python are designed for the efficient processing of large datasets by performing operations lazily:
Here, the read_csv function in Polars is used to create a DataFrame representing the data in the CSV file, but the data isn't loaded into RAM until processing is triggered.
We define a computation (sum of some_column), and only when we call collect(), the actual computation is executed.
The result of collect() is another DataFrame, from which we extract the value of the sum.
Data structures
The right data structure (e.g., lists, dictionaries, sets) can drastically improve an operation's memory and computational efficiency.
Suppose you are working on a data engineering task where you need to check whether certain items exist in a dataset. Let's assume you have a list of user IDs, and you need to check if specific user IDs exist in this list.
Using a list for this task is straightforward, but it's not efficient because checking for existence in a list requires scanning through the list, which can be very slow if the list is long:
A more efficient approach would be to use a set, which is designed for fast membership tests. Converting the list to a set allows for much quicker lookups:
List comprehension and generator expressions
List comprehension in Python offers a concise and efficient way to create lists, improving execution speed and code readability. Unlike traditional for-loops, list comprehensions are generally faster because they are optimized for the Python interpreter to execute a predictable pattern of accessing and processing list elements. This optimization leads to quicker list creation and manipulation.
Moreover, list comprehensions reduce the overhead of repeatedly calling an append method in a loop, as the entire list is constructed in a single, readable line. This not only makes the code more elegant but also allows it to run more efficiently by minimizing function calls and RAM usage. By handling list operations in a more streamlined fashion, Python's comprehensions can lead to performance gains, especially in scenarios involving large datasets or complex list-processing tasks.
We can run a simple example to demonstrate:
### Using a standard for-loop
squares_loop = []
for number in range(1, 1000001):
squares_loop.append(number ** 2)
### Using list comprehension
squares_list_comp = [number ** 2 for number in range(1, 1000001)]
The performance difference between a for-loop and a list comprehension might appear minimal, especially in simple operations like the one in the example. However, for more complex list processing tasks, especially those involving multiple operations within each iteration, the performance improvement can be more noticeable.
Generator object and expressions
Python generator expressions are a concise way to generate values on the fly, similar to list comprehensions, but for generating iterators.
They provide RAM efficiency, lazy evaluation (they only compute the next value when it's needed), and are most useful when working with large data sets where you want to process items one at a time, or when you only need a part of the generated sequence, thus avoiding the overhead of generating the complete list.
Generator expressions can be easily used with built-in functions like sum(), max(), min(), etc., which take iterables as inputs.
A simple example would be:
gen_exp = (x * 2 for x in range(5))
for value in gen_exp:
print(value)which will print the doubled values of 0 through 4, one at a time, without ever creating a full list in memory.
In-memory vs. compute engines
Performing computations in-memory can be more efficient for smaller tasks, but leveraging a compute engine becomes essential for data-intensive tasks.
Suppose you are tasked with aggregating a large dataset to compute statistics like mean, sum, and count per group.
For smaller datasets, you might use a library like Pandas to perform these computations in-memory.
For larger datasets or more complex operations, you might leverage a compute engine like Apache Spark, which can handle distributed data processing across multiple machines.
Beyond standalone compute engines like Apache Spark, databases and data warehouses also act as powerful compute engines. Modern data warehouses, such as Snowflake, BigQuery, or Redshift, are designed to run complex analytical queries directly on large datasets.
Instead of pulling data out into a separate compute environment, you can use the computational capabilities of these systems directly, which reduces the need to transfer data and have additional processing layers.
In conclusion: optimizing data pipeline Python code
At its core, data orchestration is about coordinating various data operations into cohesive workflows. Processes can become agonizingly slow or falter under the weight of poorly optimized Python code, crippling analytics and hampering critical decision-making.
As you progress in your programming career, aim to go beyond building pipelines that work and architect efficient and robust data solutions optimized for performance from day one.